Overview of the FarmGTEx Project
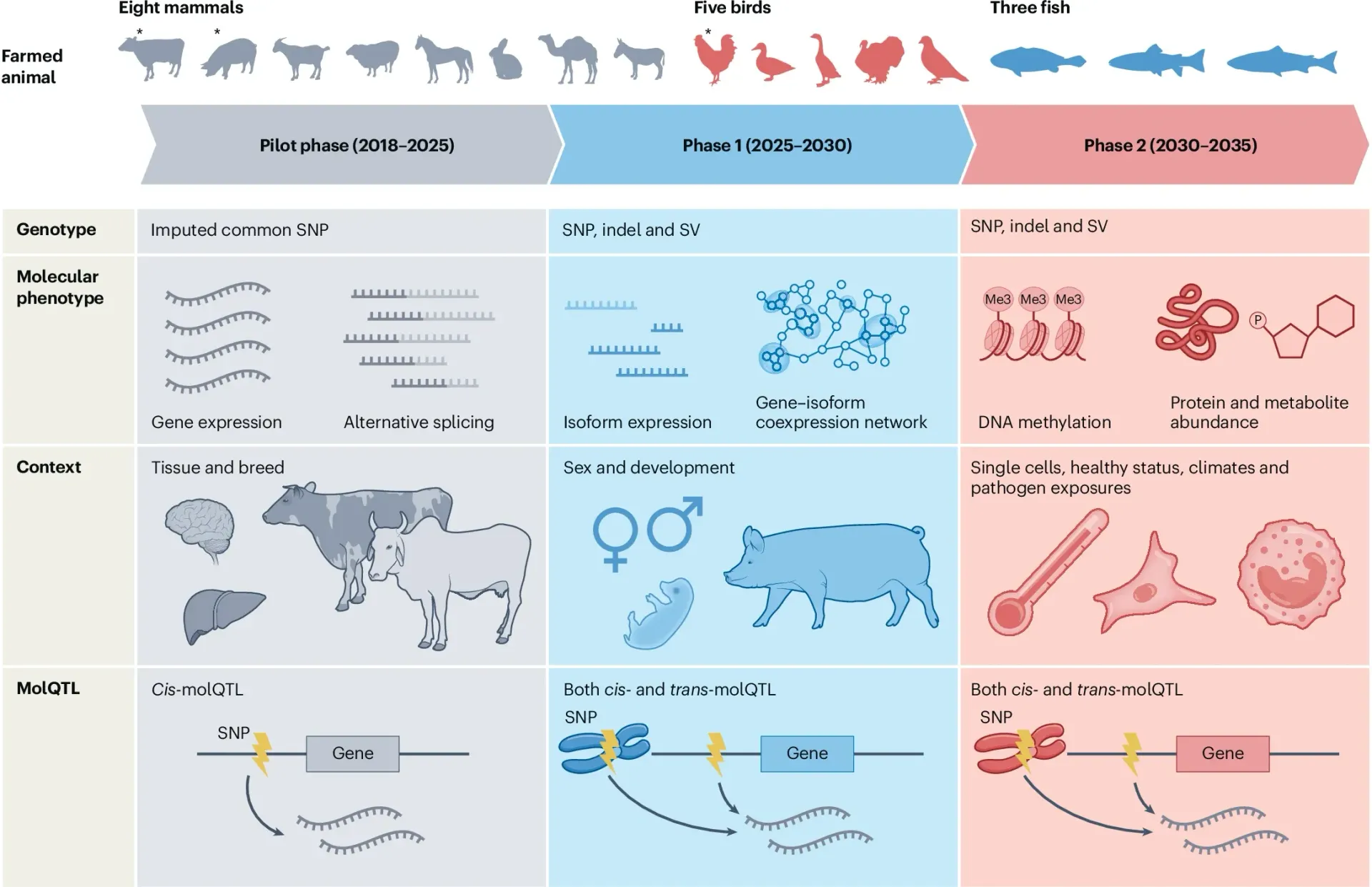
To date, we are considering 16 farmed animal species, including eight mammals (that is, cattle, pigs, goats, sheep, horses, rabbits, camels and donkeys), five birds (that is, chickens, ducks, geese, turkeys and pigeons) and three fish (that is, large yellow croaker, Atlantic salmon and rainbow trout). Additional farmed species will be considered as the project progresses. The regulatory effects of genomic variants on different molecular phenotypes across biological contexts and environmental conditions will be systematically exploited using cis-molQTL and trans-molQTL mapping. Different goals and milestones for the three phases of FarmGTEx are outlined. Asterisks indicate that the pilot phases for these species have been completed. Indel, short insertion and deletion (length less than 50 nucleotides); SNP, single nucleotide polymorphism; SV, structural variants.
The overall data analysis pipeline of FarmGTEx
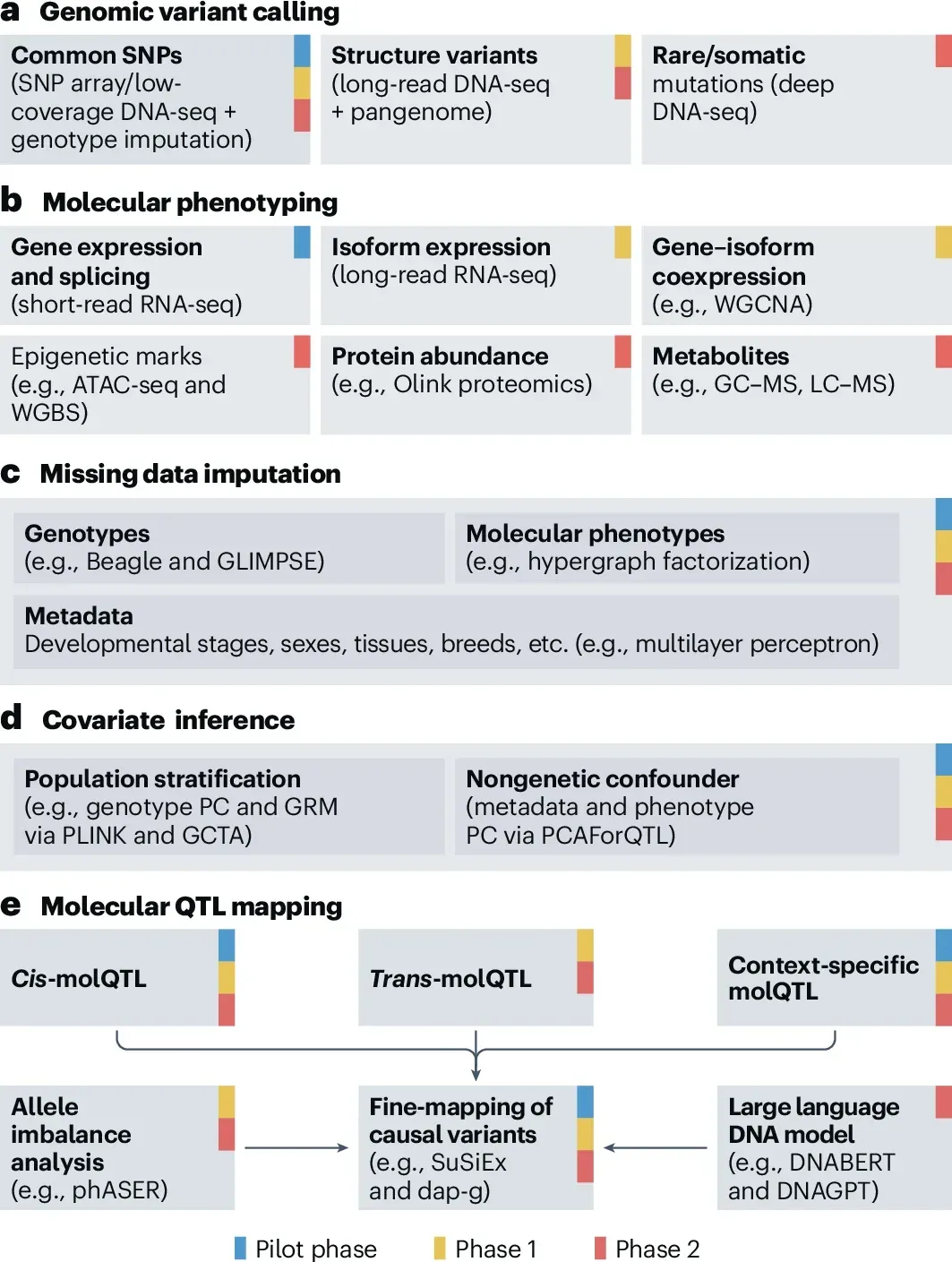
a–e, The flowchart highlights the key stages and methodologies of data generation and analyses across the three phases of FarmGTEx, including genomic variant calling (a), definition and quantification of molecular phenotypes (b), missing data imputation (c), batch effect inference and removal (d), and molQTL mapping via, for example, OmiGA (https://omiga.farmgtex.org) and ClipperQTL (e). Examples of approaches and methods used for each stage are shown in parentheses. ATAC-seq, assay for transposase-accessible chromatin with sequencing; dap-g, deterministic approximation of posteriors: genetics; DNABERT; DNAGPT; DNA-seq, DNA sequencing; GC–MS, gas chromatography–mass spectrometry; GCTA, a tool for genome-wide complex trait analysis; GRM, genetic relationship matrix; GLIMPSE, genotype likelihoods imputation and phasing method; LC–MS, liquid chromatography–mass spectrometry; PC, principal component; PLINK, a tool for whole-genome association and linkage analyses; phASER, phasing and allele-specific expression from RNA-seq; SuSiEx, an enhanced tool for cross-ancestry fine-mapping of causal variants based on ‘sum of single effects’; WGBS, whole-genome bisulfite sequencing; WGCNA, weighted gene coexpression network analysis.
Potential applications of FarmGTEx resources

Insights into the dynamic landscape of the regulatory effects of genomic variants across diverse biological contexts (for example, sex and developmental stage) can advance our understanding of how genetics, development and sex interact with complex phenotypes. Identified causal variants and genes for complex traits and diseases can be used in precision animal breeding via gene editing and synthetic biology. Determining the genetic control of molecular phenotypes in global farmed animal breeds and their wild progenitors and congeners can enable us to understand the molecular mechanisms that underlie domestication and local environmental adaptation. Comparative analyses of farmed animals and humans across multiple biological layers can help to develop suitable animal models for studying human biology, disease and xenotransplantation.








